ANNOVAR (3): 更新 COSMIC 数据库 (v70+)
由于 COSMIC 更新了许可限制,因此 ANNOVAR 提供的 COSMIC 数据库注释最后一版是 v70,之后的版本虽然 ANNOVAR 没有提供,但是 Kai Wang 却很贴心地提供了更新的脚本,用户可以根据文档,自己将 v70之后版本的 COSMIC 转换成 ANNOVAR 的注释数据库。
所需原始数据及程序
COSMIC 数据库 V83 后,以下文件可以直接通过官网点击 Data -> Downloads 下载,不需要再使用 SFTP 下载了。
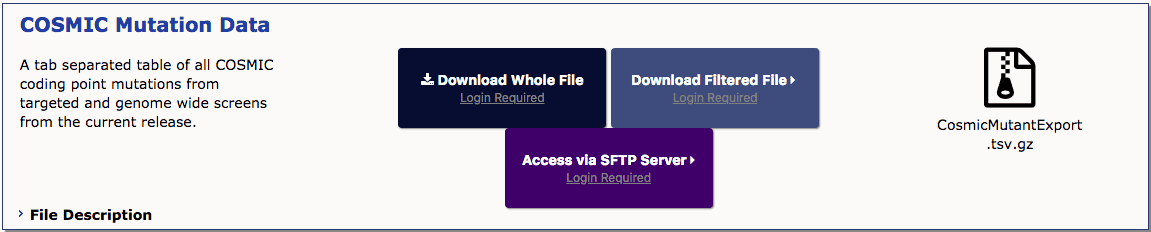
- COSMIC的VCF文件,可以分为 Coding Variant 或者 Non Coding Variant 两种,如
CosmicCodingMuts.vcf.gz和CosmicNonCodingVariants.vcf.gz - COSMIC MutantExport file,也可以分为 Coding Variant 和 Non Coding Variant 两种,如
CosmicMutantExport.tsv.gz和CosmicNCV.tsv - ANNOVAR的转换脚本,如
prepare_annovar_user.pl - ANNOVAR数据库的index脚本 Annovar_index.pl (该脚本在官方文档里面是没有的,by 张求学&周在威)
处理过程
prepare_annovar_user.pl -dbtype cosmic CosmicMutantExport.tsv -vcf CosmicCodingMuts.vcf > hg38_cosmic81_coding.txt # 生成 Coding Variant 的注释文件
prepare_annovar_user.pl -dbtype cosmic CosmicNCV.tsv -vcf CosmicNonCodingVariants.vcf > hg38_cosmic81_noncoding.txt # 生成 Non Coding Variant 的注释文件
## 以下步骤是我自行添加的,可以忽略 ##
sort -k1 -V -s -t ' ' hg38_cosmic81_coding.txt > hg38_cosmic81_coding.sorted.txt #排序
perl Annovar_index.pl hg38_cosmic81_coding.sorted.txt 1000 #生成index,但其实注释文件很小,也可以不生成
mv hg38_cosmic81_coding.sorted.txt hg38_cosmic81_coding.txt
mv hg38_cosmic81_coding.sorted.txt.idx hg38_cosmic81_coding.txt.idxANNOVAR 原文引用
Note that the prepare_annovar_user.pl file can be downloaded from here. The final result file should contain coding mutations from COSMIC, as well as the number of occurrences in different tumor types (However, note that these include both targeted screen and genome screen. If you only want genome screen, you should use the CosmicGenomeScreensMutantExport.tsv.gz file instead).
Recently, COSMIC changed their data formats so non-coding mutations are no longer in the MutantExport file, so we can no longer calculate their occurrences in various tumors. COSMIC now provides a CosmicNCV.tsv file, but it is not really that informative as the cancer tissue information is missing from this file.
However, as of 2017, in more recent versions of COSMIC, the noncoding variants are now included in CosmicNCV.tsv file, so that we can use this file to annotate noncoding variants. In early 2017, the prepare_annovar_user.pl script was updated to handle noncoding variants in COSMIC. An example is given below for cosmic81:
prepare_annovar_user.pl -dbtype cosmic CosmicMutantExport.tsv -vcf CosmicCodingMuts.vcf > hg38_cosmic81_coding.txt
prepare_annovar.pl -dbtype cosmic CosmicNCV.tsv -vcf CosmicNonCodingVariants.vcf > hg38_cosmic81_noncoding.txtThere should be 2.58M coding and 14.2M noncoding variants, after you run the commands above. Users cannot index the file, but the file size is not too large, and you do not need to use indexing to use ANNOVAR.
